Other topics
This module has concentrated on some topics in numerical analysis and numerical computation that are quite important for statistics. It has not covered a great many other topics in numerical analysis that are also important. Despite its name the module has made no attempt whatsoever to provide an overview of ‘statistical computing’ more generally, which would surely have to talk about data handling, pattern recognition, sorting, matching, searching, algorithms for graphs and trees, computational geometry, discrete optimization, the EM algorithm, dynamic programming, linear programming, fast Fourier transforms, wavelets, support vector machines, function approximation, sparse matrices, huge data sets, graphical processing units, multi-core processing and a host of other topics.
Rather than attempt the impossible task of trying to remedy this deficiency, we will look at some examples of other topics.
1 Random number generation
The stochastic integration methods of the last section took it for granted that we can produce random numbers from various distributions. Actually, we can’t. The best that can be done is to produce a completely deterministic sequence of numbers which appears indistinguishable from a random sequence with respect to any relevant statistical property that we choose to test. In other words, we may be able to produce a deterministic sequence of numbers that can be very well modelled as being a random sequence from some distribution. Such deterministic sequences are referred to as sequences of pseudorandom numbers, but the pseudo part usually gets dropped at some point.
The fundamental problem, for our purposes, is to generate a pseudorandom sequence that can be extremely well modelled as a sequence of realizations of i.i.d. \(U(0,1)\) random variables. Given such a sequence, it can be straightforward to generate deviates from other distributions, but the i.i.d. \(U(0,1)\) generation is where one problem lies. Indeed if you read around this topic then most books will pretty much agree about how to turn uniform random deviates into deviates from a huge range of other distributions, but advice on how to get the uniform deviates in the first place is much less consistent.
1.1 Simple generators and what can go wrong
Since the 1950s there has been much work on linear congruential generators. The intuitive motivation is something like this. Suppose I take an integer, multiply it by some enormous factor, re-write it in base - ‘something huge’, and then throw away everything except for the digits after the decimal point. Pretty hard to predict the result, no? So, if I repeat the operation, feeding each step’s output into the input for the next step, a more or less random sequence might result. Formally, the pseudorandom sequence is defined by \[ X_{i+1} = a X_i + b ~ ({\rm mod} ~M) \] where \(b\) is 0 or 1, in practice. This is started with a seed \(X_0\). The \(X_i\) are integers (\(<M\), of course), but we can define \(U_i = X_i/M\). Now the intuitive hope that this recipe might lead to \(U_i\) that are reasonably well modelled by i.i.d. \(U(0,1)\) r.v.s is only realised for some quite special choices of \(a\) and \(M\), and it takes some number theory to give the generator any sort of theoretical foundation (see Ripley, 1987, Chapter 2).
An obvious property to try to achieve is full period. We would like the generator to visit all possible integers between \(1-b\) and \(M-1\) once before it starts to repeat itself (clearly the first time it revisits a value, it starts to repeat itself). We would also like successive \(U_i\)s to appear uncorrelated. A notorious and widely used generator called RANDU, supplied at one time with IBM machines, met these basic considerations with \[ X_{i+1} = 65539 X_i ~ ({\rm mod} ~ 2^{31}) \] This appears to do very well in 1 dimension.
n <- 100000 ## code NOT for serious use
x <- rep(1,n)
a <- 65539;M <- 2^31;b <- 0 ## Randu
for (i in 2:n) x[i] <- (a*x[i-1]+b)%%M
u <- x/(M-1)
qqplot((1:n-.5)/n,sort(u))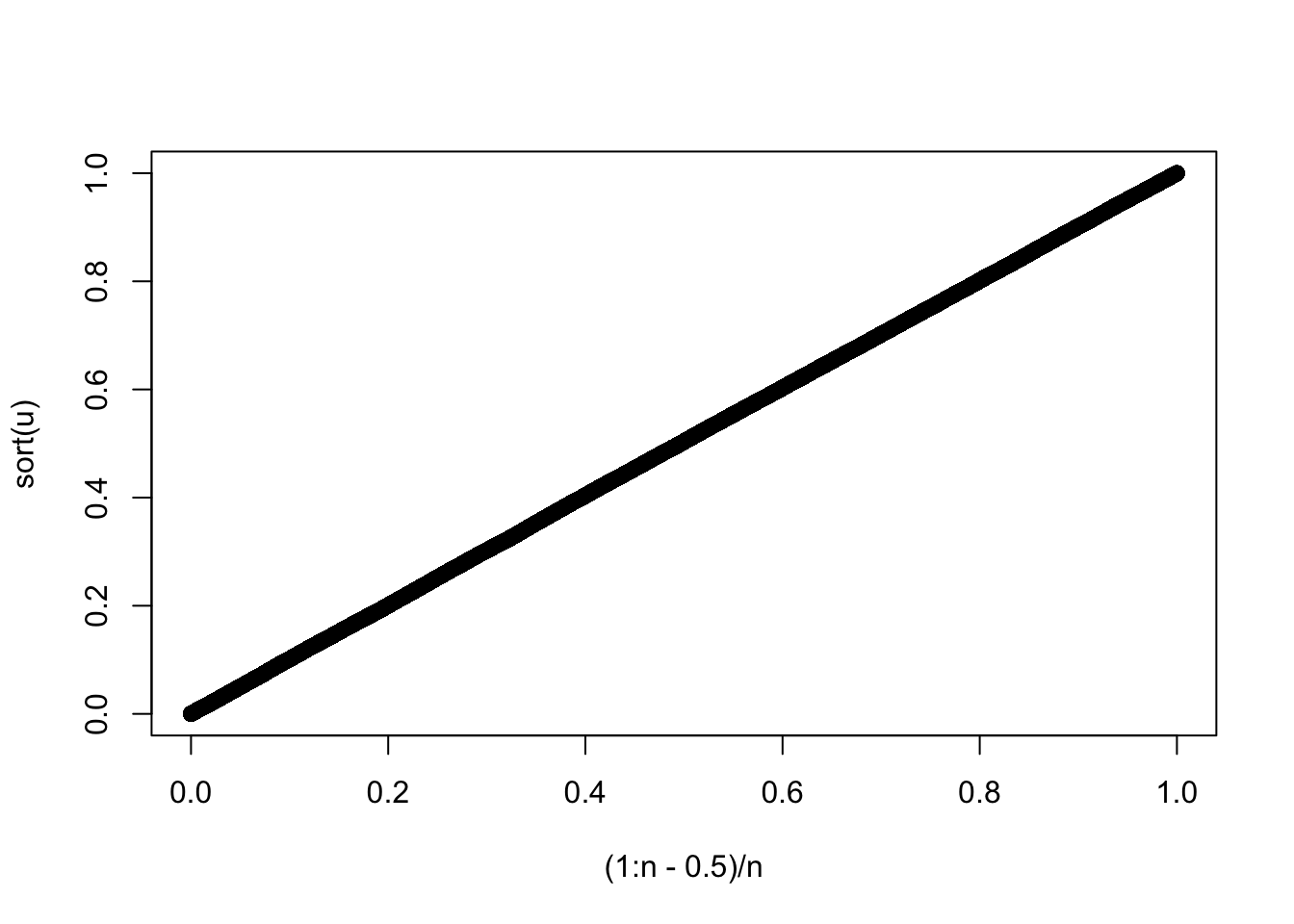
Similarly a 2D plot of \(U_i\) vs \(U_{i-1}\) indicates no worries with serial correlation
## Create data frame with U at 3 lags...
U <- data.frame(u1=u[1:(n-2)],u2=u[2:(n-1)],u3=u[3:n])
plot(U$u1,U$u2,pch=".")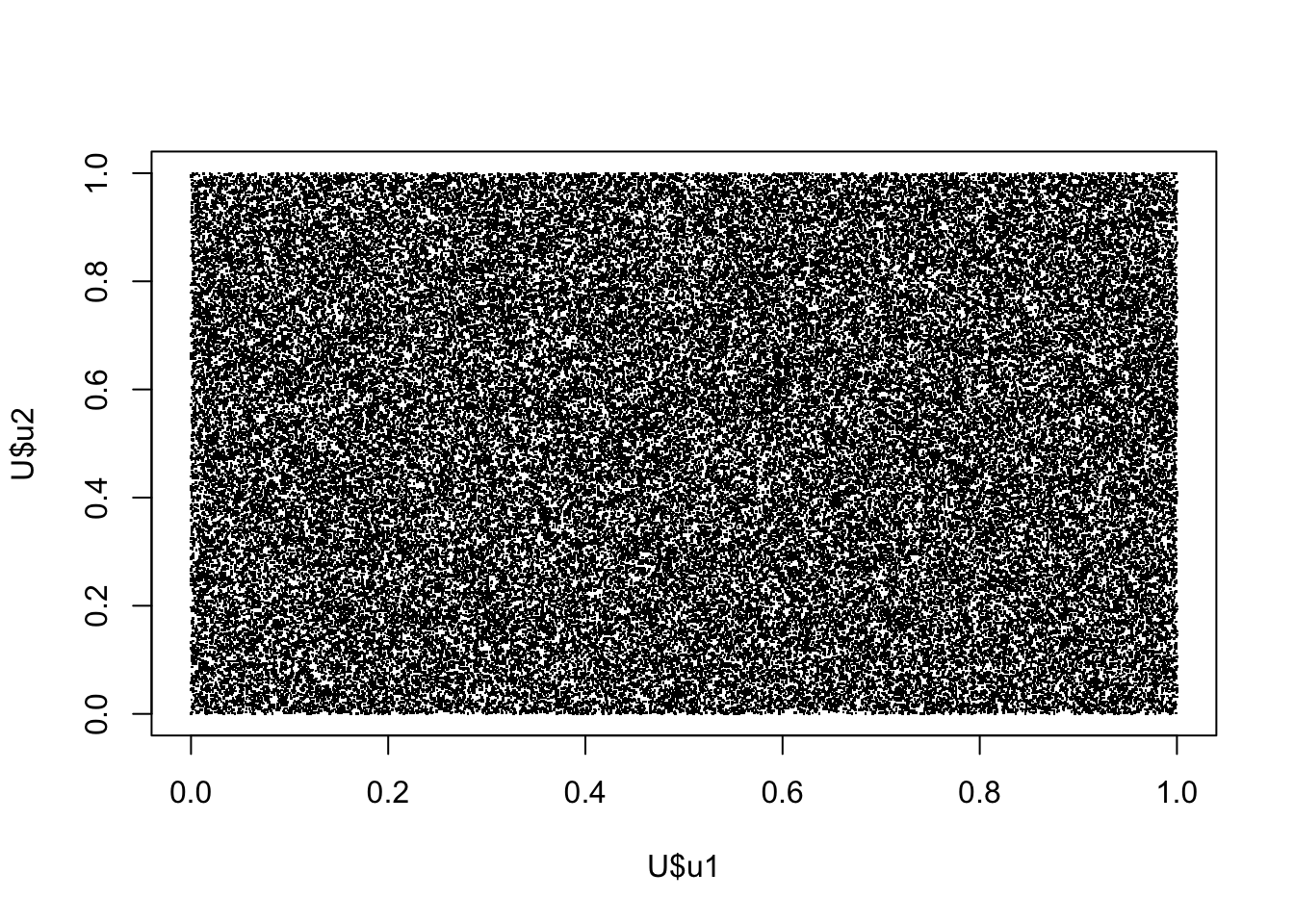
We can also check visually what the distribution of the triples \((U_i, U_{i-1},U_{i-2})\) looks like.
library(lattice)
cloud(u1~u2*u3,U,pch=".",col=1,screen=list(z=40,x=-70,y=0))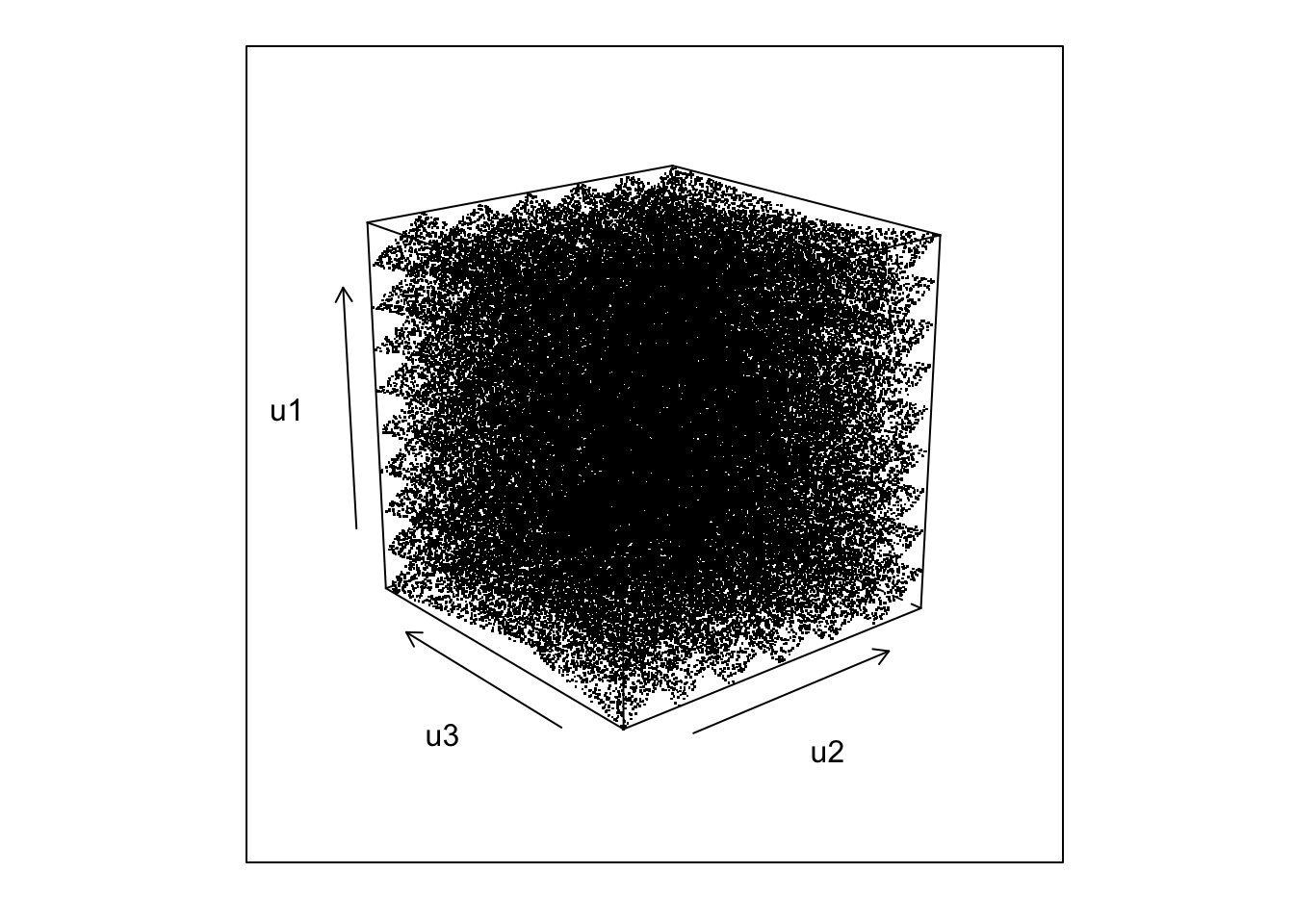
not quite so random looking. Experimenting a little with rotations gives
cloud(u1~u2*u3,U,pch=".",col=1,screen=list(z=40,x=70,y=0))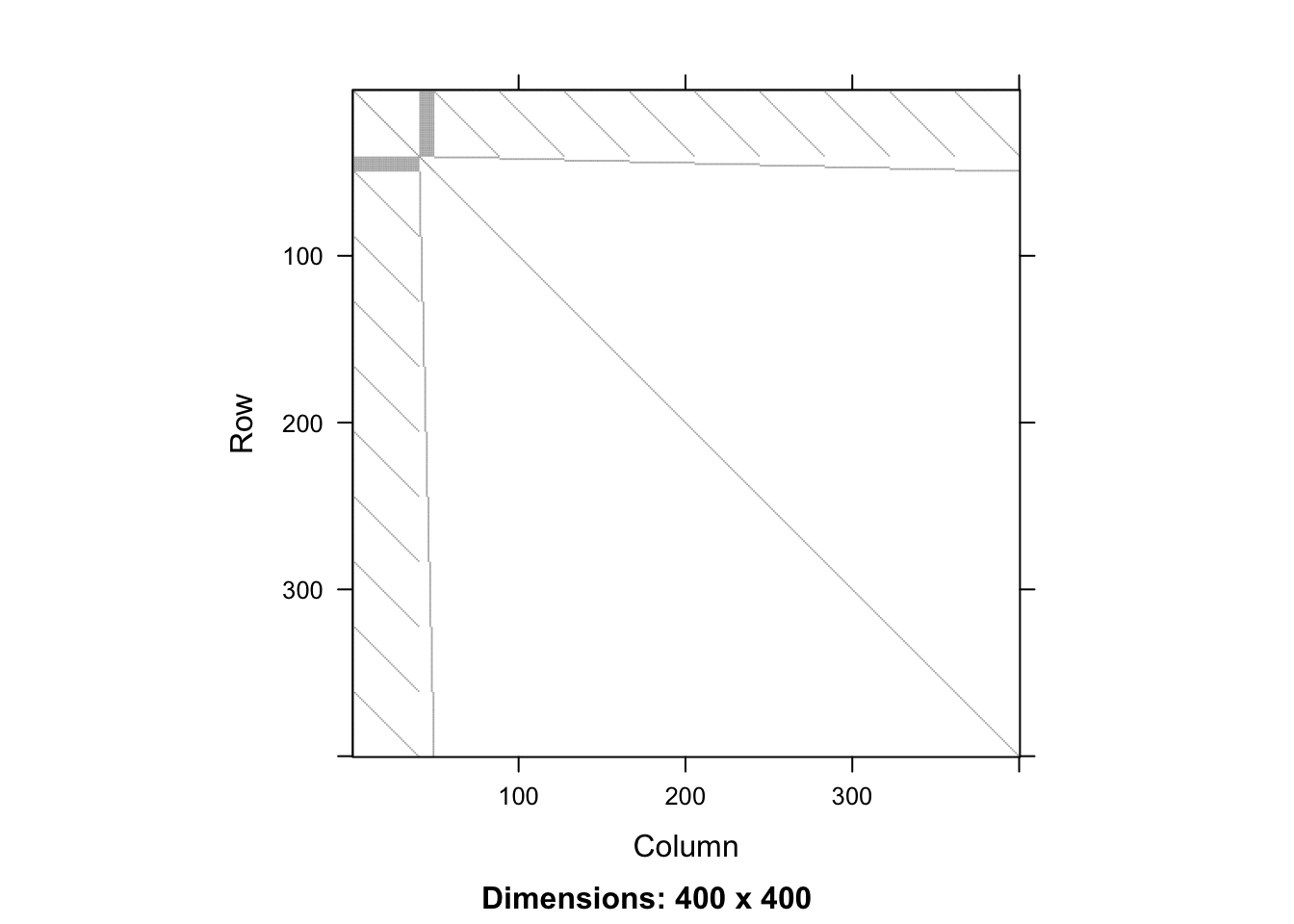
i.e. the triples lie on one of 15 planes. Actually you can show that this must happen (see Ripley, 1987, section 2.2).
Does this deficiency matter in practice? When we looked at numerical integration we saw that quadrature rules based on evaluating integrands on fixed lattices become increasingly problematic as the dimension of the integral increases. Stochastic integration overcomes the problems by avoiding the biases associated with fixed lattices. We should expect problems if we are performing stochastic integration on the basis of random numbers that are in fact sampled from some coarse lattice.
So the first lesson is to use generators that have been carefully engineered by people with a good understanding of number theory, and have then been empirically tested (Marsaglia’s Diehard battery of tests provides one standard test set). For example, if we stick with simple congruential generators, then \[ X_i = 69069 X_{i-1}+1 ~ ({\rm mod} ~2^{32}) \tag{1}\] is a much better bet. Here is its triples plot. No amount of rotating provides any evidence of structure.
Although this generator is much better than RANDU, it is still problematic. An obvious infelicity is the fact that a very small \(X_i\) will always be followed by an unusually small \(X_{i+1}\) (consider \(X_i = 1\), for example). This is not a property that would be desirable in a time series simulation, for example. Not quite so obvious is the fact that for any congruential generator of period \(M\), then \(k-\)tuples, \(U_i, U_{i-1}, \ldots, U_{i-k+1}\) will tend to lie on a finite number of \(k-1\) dimensional planes (e.g. for RANDU we saw 3-tuples lying on 2 dimensional planes.) There will be at most \(M^{1/k}\) such planes, and as RANDU shows, there can be far fewer. The upshot of this is that if we could visualise 8 dimensions, then the 8-tuple plot for Equation 1 would be just as alarming as the 3D plot was for RANDU. 8 is not an unreasonably large dimension for an integral.
Generally then, it might be nice to have generators with better behaviour than simple congruential generators, and in particular we would like generators where \(k-\)tuples appear uniformly distributed on \([0,1]^k\) for as high a \(k\) as possible (referred to as having a high k-distribution}).
1.2 Building better generators
An alternative to the congruential generators are generators which focus on generating random sequences of 0s and 1s. In some ways this seems to be a natural representation of numbers when using modern digital computers. One family of generators are those often termed shift-register generators. The basic approach is to use bitwise binary operations to make a binary sequence `scramble itself’. An example is Marsaglia’s (2003) Xorshift generator as recommended in Press et al. (2007).
Let \(x\) be a 64-bit variable (i.e. an array of 64 0s or 1s). The generator is initialised by setting to any value (other than 64 0s). The following steps then constitute one iteration (update of \(x\)) \[\begin{eqnarray*} x &\leftarrow& x \wedge (x \gg a) \\ x &\leftarrow& x \wedge (x \ll b) \\ x &\leftarrow& x \wedge (x \gg c) \end{eqnarray*}\] each iteration generates a new random sequence of 0s and 1s. \(\wedge\) denotes `exclusive or’ (XOR) and \(\gg\) and \(\ll\) are right-shift and left shift respectively, with the integers \(a, b\) and \(c\) giving the distance to shift. \(a=21\), \(b=35\) and \(c=4\) appear to be good constants (but see Press et al., 2007, for some others).
If you are a bit rusty on these binary operators then consider an 8-bit example where x=10011011 and z=01011100.
x<<1is00110110, i.e. the bit pattern is shifted leftwards, with the leftmost bit discarded, and the rightmost set to zero.x<<2is01101100, i.e. the pattern is shifted 2 bits leftwards, which also entails discarding the 2 leftmost bits and zeroing the two rightmost.x>>1is01001101, same idea, but rightwards shift.x^zis11000111, i.e. a 1 where the bits inxandzdisagree, and a 0 where they agree.
The Xorshift generator is very fast, has a period of \(2^{64}-1\), and passes the Diehard battery of tests (perhaps unsurprising as Marsaglia is responsible for that too). These shift register generators suffer similar granularity problems to congruential generators (there is always some \(k\) for which \([0,1]^k\) can not be very well covered by even \(2^{64}-1\) points), but tend to have all bit positions `equally random’, whereas lower order bits from congruential generator sequences often have a good deal of structure.
Now we reach a fork in the road. To achieve better performance in terms of longer period, larger k-distribution, and fewer low order correlation problems, there seem to be two main approaches, the first pragmatic, and the second more theoretical.
Combine the output from several ‘good’, well understood, simple generators using operations that maintain randomness (e.g. XOR and addition, but not multiplication). When doing this, the output from the combined generators is never fed back into the driving generators. Preferably combine rather different types of generator. Press et al. (2007) make a convincing case for this approach. Wichmann-Hill, available in R is an example of such a combined generator, albeit based on 3 very closely related generators.
Use more complicated generators — non-linear or with a higher dimensional state that just a single \(X_i\) (see Gentle, 2003). For example, use a shift register type generator based on maintaining the history of the last \(n\) bit-patterns, and using these in the bit scrambling operation. Matsumoto and Nishimura’s (1998) Mersenne Twister is of this type. It achieves a period of \(2^{19937}-1\) (that’s not a misprint: \(2^{19937}-1\) is a ‘Mersenne prime’ (numbers this large are often described as being `astronomical’, but this doesn’t really do it justice: there are probably fewer than \(2^{270}\) atoms in the universe), and is 623-distributed at 32 bit accuracy. i.e. its 623-tuples appear uniformly distributed (each appearing the same number of times in a full period) and spaced \(2^{-32}\) apart (without the ludicrous period this would not be possible). It passes Diehard, is the default generator in R, and C source code is freely available.
1.3 Uniform generation conclusions
I hope that the above has convinced you that random number generation, and use of pseudo-random numbers are non-trivial topics that require some care. That said, most of the time, provided you pick a good modern generator, you will probably have no problems. As general guidelines
Avoid using black-box routines supplied with low level languages such as C, you don’t know what you are getting, and there is a long history of these being botched.
Do make sure you know what method is being used to generate any uniform random deviates that you use, and that you are satisfied that it is good enough for purpose.
For any random number generation task that relies on \(k-\)tuples having uniform distribution for high \(k\) then you should be particularly careful about what generator you use. This includes any statistical task that is somehow equivalent to high dimensional integration.
The Mersenne Twister is probably the sensible default choice in most cases, at present. For high dimensional problems it remains a good idea to check answers with a different high-quality generator. If results differ significantly, then you will need to find out why (probably starting with the `other’ generator).
Note that I haven’t discussed methods used by cryptographers. Cryptographers want to use (pseudo)random sequences of bits (0s and 1s) to scramble messages. Their main concern is that if someone were to intercept the random sequence, and guess the generator being used, they should not be able to infer the state of the generator. Achieving this goal is quite computer intensive, which is why generators used for cryptography are usually over engineered for simulation purposes.
1.4 Other deviates
Once you have a satisfactory stream of i.i.d. \(U(0,1)\) deviates then generating deviates from other standard distributions is much more straightforward. Conceptually, the simplest approach is inversion. We know that if \(X\) is from a distribution with continuous c.d.f. \(F\) then \(F(X)\sim U(0,1)\). Similarly, if we define the inverse of \(F\) by \(F^-(u) = {\rm min}(x|F(x)\ge u)\), and if \(U\sim U(0,1)\), then \(F^-(U)\) has a distribution with c.d.f. \(F\) (this time with not even a continuity restriction on \(F\) itself).
As an example here is inversion used to generate 1 million i.i.d. \(N(0,1)\) deviates in R.
system.time(X <- qnorm(runif(1e6))) user system elapsed
0.019 0.001 0.020 For most standard distributions (except the exponential) there are better methods than inversion, and the happy situation exists where textbooks tend to agree what these are. Ripley’s (1987) Chapter 3 is a good place to start, while the lighter version is provided by Press et al. (2007) Chapter 7. R has many of these methods built in. For example an alternative generation of 1 million i.i.d. \(N(0,1)\) deviates would use.
system.time(Z <- rnorm(1e6)) user system elapsed
0.026 0.001 0.027 Here the difference is marginal, due to the highly optimized qnorm function in R, but for other distributions the difference can be substantial.
We have already seen how to generate multivariate normal deviates given i.i.d. \(N(0,1)\) deviates, and for other (simple) multivariate distributions see chapter 4 of Ripley (1987). For more complicated distributions see the Computationally-Intensive Statistical Methods APTS module.
2 Sparse matrix computation
Many statistical computations involve matrices which contain very high proportions of zeroes: these are known as sparse matrices. Most of the computational time used by most numerical linear algebra routines is devoted to additions, subtractions and multiplications of pairs of floating point numbers: if we know, in advance, that one of the pair is a zero, we don’t actually need to perform the operation, since the answer is also known in advance. In addition, there is little point in storing all the zeroes in a sparse matrix: it is often much more efficient to store only the values of the non-zero elements, together with some encoding of their location in the matrix.
2.1 Example: a design matrix with categorical variables
Efficient storage and simple matrix manipulations are easy to implement. Here is an example, based on simulating the model matrix for a linear model with 2 factors (i.e. with 2 categorical variables), and a large number of observations.
n <- 100000 ## number of obs
pa <- 40;pb <- 10 # numbers of factor levels
a <- factor(sample(1:pa,n,replace=TRUE))
b <- factor(sample(1:pb,n,replace=TRUE))
X <- model.matrix(~a*b) # model matrix a + b + a:b
y <- rnorm(n) # a random response
object.size(X) # X takes lots of memory!326428560 bytes sum(X==0)/prod(dim(X)) # proportion of zeros[1] 0.9906331The simulated \(\bf X\) is \(100,000 \times 400\) and is about 99% zeroes. It requires about 0.3Gb of computer memory, if stored as a normal ‘dense’ matrix. The Matrix library, supplied with R, supports sparse matrix computations, so the following produces a sparse version of \(\bf X\).
par(mar=c(4,4,1,1))
library(Matrix)
Xs <- Matrix(X) ## a sparse version of X
## much reduced memory footprint
as.numeric(object.size(Xs))/as.numeric(object.size(X))[1] 0.03346803Because Xs doesn’t store the zeroes, it uses much less memory than the dense version X. Note that although X is only 1% non-zeroes, Xs uses 3% of the storage of X, because it needs to store the locations of the non-zero elements, in addition to their values. The sparse approach also saves computer time when performing basic matrix operations, by avoiding the floating point operations that are not needed because they involve a zero. For example
system.time(Xy <- t(X)%*%y) user system elapsed
0.096 0.027 0.123 system.time(Xsy <- t(Xs)%*%y) user system elapsed
0.003 0.001 0.004 Again computational overheads involved in only storing the non-zeroes, and their locations, means that the cost of the sparse operation is more than 1% of the dense cost, but the saving is still substantial.
Any competent programmer could produce code for the examples given so far, but serious applications may involve solving linear systems, and this is much less straightforward. Sparse versions of some of the decompositions that we met before are needed, but naive implementations using sparse matrices flounder on the problem of infill (or fill-in). You can see the problem in the following dense calculation
system.time({ XX <- t(X)%*%X; R <- chol(XX) }) user system elapsed
0.221 0.026 0.181 dim(XX)[1] 400 400sum(XX == 0)/prod(dim(XX))[1] 0.97935image(as(XX,"Matrix"))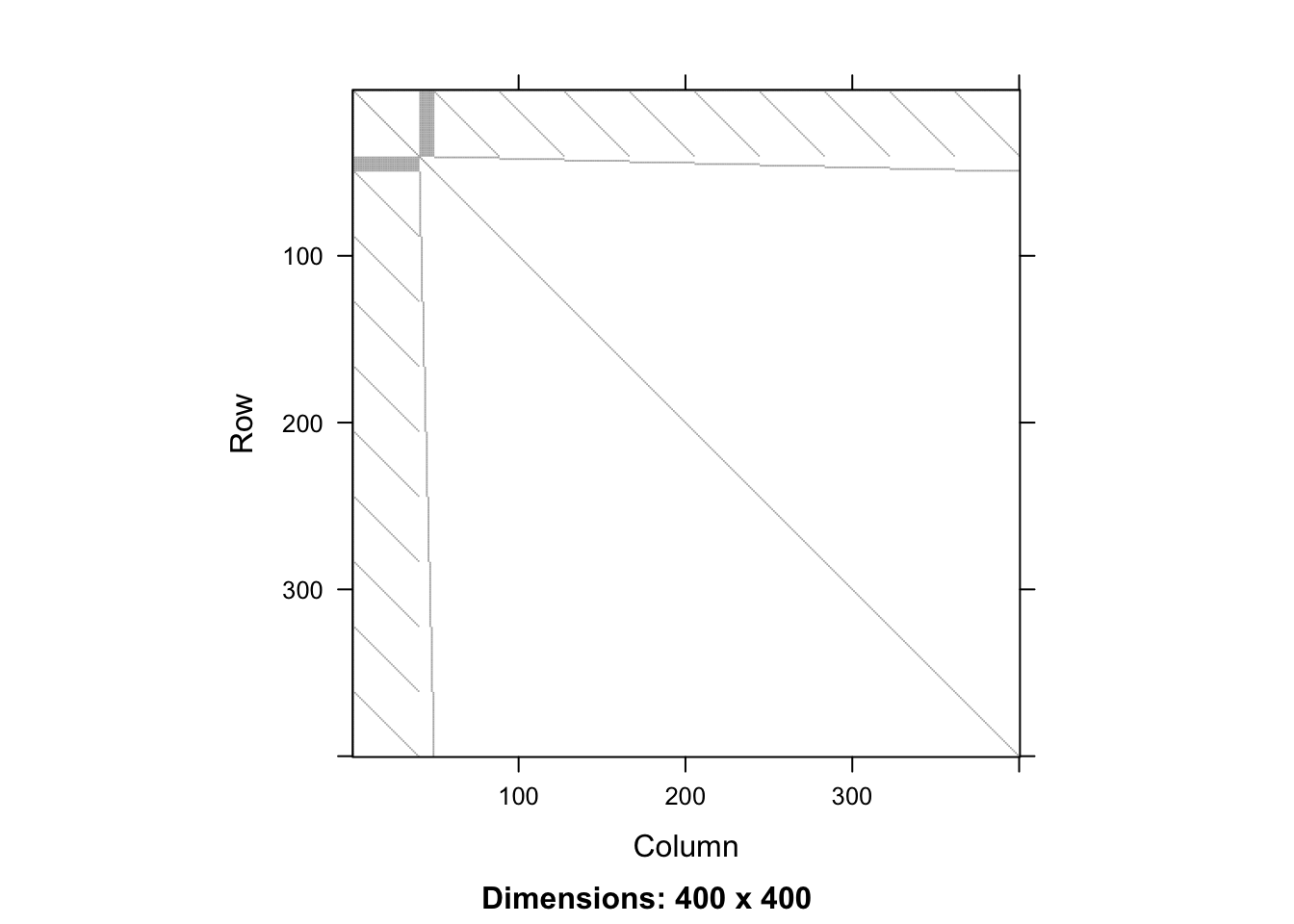
range(t(R)%*%R-XX)[1] -1.818989e-12 1.455192e-11sum(R != 0) ## the upper triangle is dense (not good)[1] 80200image(as(R,"Matrix"))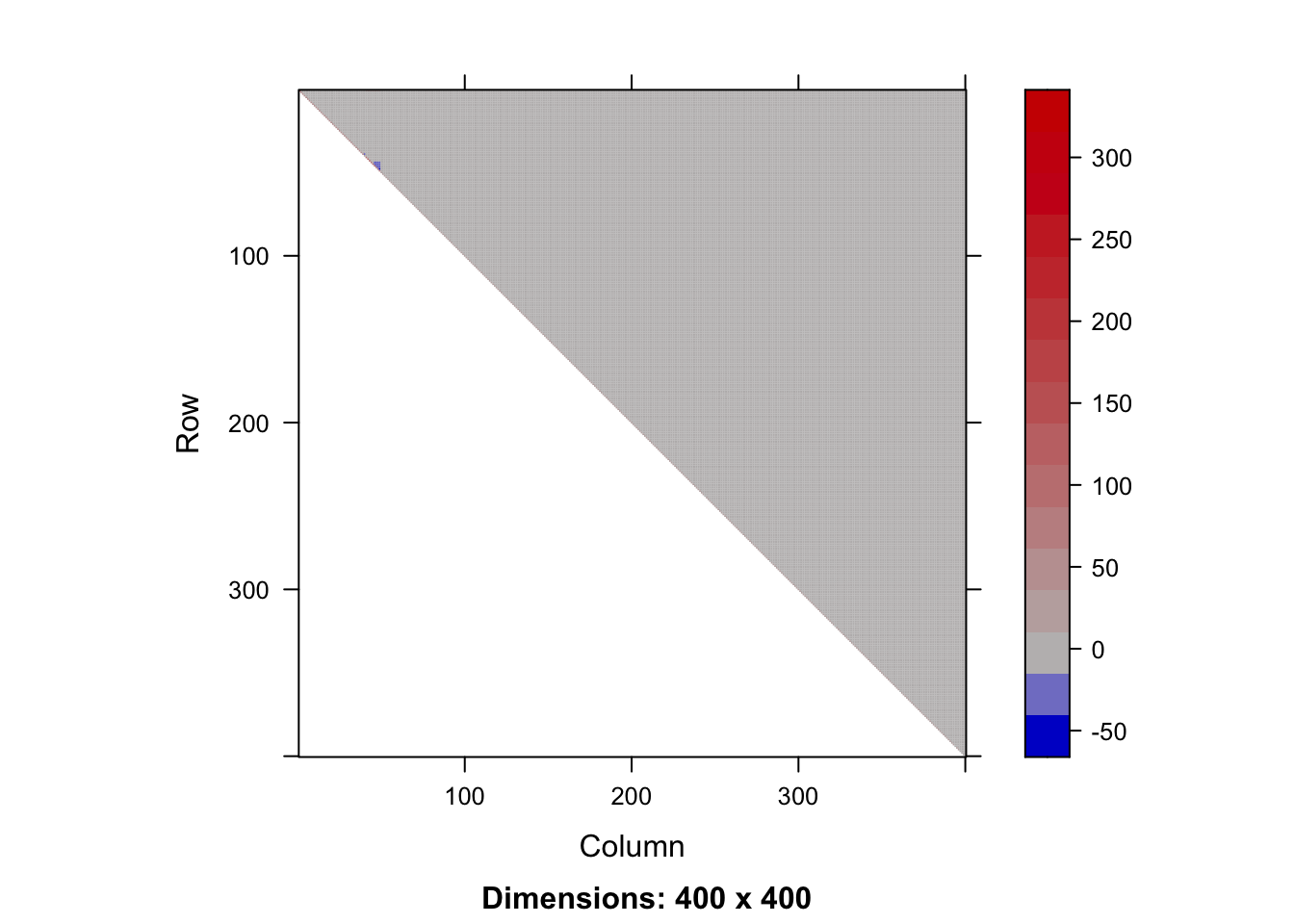
R[1:5, 1:5] ## top left corner, for example (Intercept) a2 a3 a4 a5
(Intercept) 316.2278 7.763392 7.570493 7.940479 7.633738
a2 0.0000 48.935976 -1.201012 -1.259708 -1.211046
a3 0.0000 0.000000 48.324375 -1.275263 -1.225999
a4 0.0000 0.000000 0.000000 49.444268 -1.288412
a5 0.0000 0.000000 0.000000 0.000000 48.488105Although \({\bf X}^{\sf T}{\bf X}\) is 98% zeroes, the upper triangle of its Cholesky factor is 0% zeroes, when computed in the usual way. i.e. we have lost all sparsity. Fortunately, it is possible to re-arrange the rows and columns of \({\bf X}^{\sf T}{\bf X}\) (or any other matrix for which we need a sparse Cholesky factor), in a way that reduces infill in the Cholesky factor. This pivoting means that we get a re-ordered version of the solution of the system of interest: a minor inconvenience. Unfortunately, finding the optimal pivoting in terms of infill reduction is NP hard, but there are practical computational pivoting strategies that work well in practice. These rely on some rather ingenious graph theory based symbolic analysis of the matrix to be factorised, which is applied before anything numerical is computed. See Davis (2006). Here’s what happens when a sparse Cholesky factorisation, with infill reducing pivoting, is used on \({\bf X}^{\sf T}{\bf X}\).
system.time({
XXs <- t(Xs)%*%Xs
chX <- Cholesky(XXs,perm=TRUE) ## pivoting reduces infill
Rs <- t(expand1(chX,"L")) ## extract actual factor
}) user system elapsed
0.009 0.000 0.009 piv <- chX@perm+1
range(t(Rs)%*%Rs-XXs[piv,piv]) ## factorization works![1] -5.684342e-14 4.547474e-13 sum(Rs!=0) ## the upper triangle is sparse (much better)[1] 1501 image(Rs)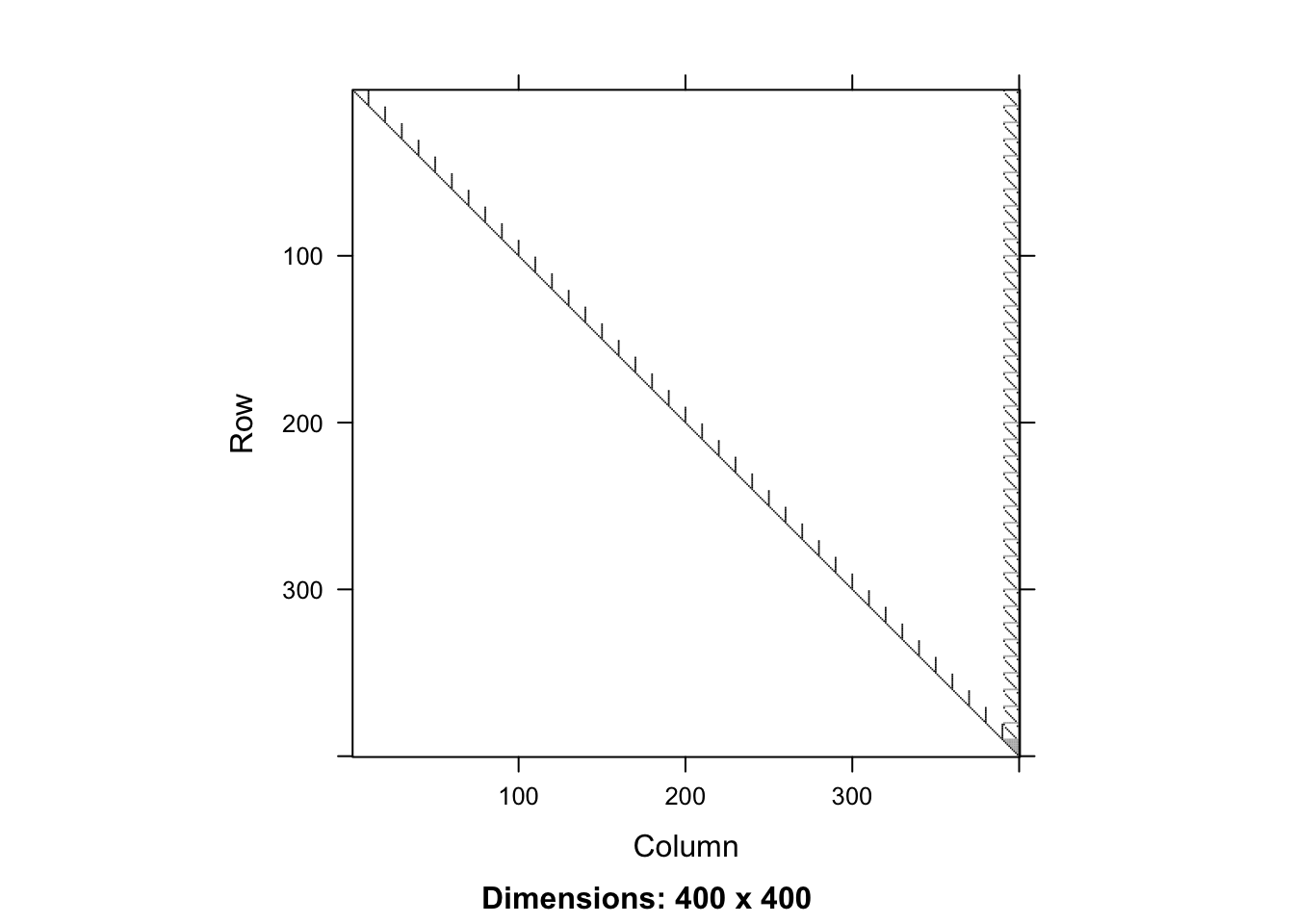
Rs[1:5,1:5] ## top left corner, for example5 x 5 sparse Matrix of class "dtCMatrix"
[1,] 15.23155 . . . .
[2,] . 15.0333 . . .
[3,] . . 15.6205 . .
[4,] . . . 14.89966 .
[5,] . . . . 15.32971With sparsity maintained in the factor, sparse backward or forward substitution is also very efficient.
Note that while the sparse Cholesky routines in the Matrix package are state-of-the-art, the QR routines are not, and carry heavy memory overheads associated with computation of \({\bf Q}\). You’ll probably need to look for sparse multi-frontal QR routines if you need sparse QR.
2.2 Example: principal eigenvector computation
In the original, idealized, PageRank algorithm for ranking webpages, Google cast the problem as answering the question: What is the average occupation time of each webpage, based on a random web surfer who
- with probability \(\alpha\) follows links uniformly at random on the page they are on?
- with probability \(1-\alpha\) samples a page from a given (possibly personal) distribution.
There is also the issue of “dangling nodes” that can be sorted out by choosing some distribution of pages to sample from.
The relevant data is the \(n \times n\) (directed) adjacency matrix \(A\) where \(A_{ij}=1\) if there is a link from page \(i\) to page \(j\) and \(0\) otherwise, and we always set \(A_{ii}=0\). \(A\) is going to be a very sparse matrix. From this we can create a substochastic matrix \(H\) (for hyperlinks!) via \[H_{ij}=\frac{A_{ij}}{\sum_{k=1}^n A_{ik}},\] if the denominator is positive and \(H_{ij}=0\) otherwise. A stochastic matrix \(S\) is then defined via \[S = H + d w^T,\] where \(d\) is a binary vector with \(d_i=1\) if and only if \(\sum_{k=1}^n A_{ik}=0\), and \(w\) is some simple distribution (like a uniform).
The matrix \(S\) is the transition matrix of the random surfer who just chooses links uniformly at random.
Now we add the possibility of choosing from a “personalization” distribution. Set \[G = \alpha S + (1-\alpha) {\bf 1}p^T,\] where \(p\) is the personalization distribution, which we assume satisfies \(p_i > 0\) for all \(i\).
Notice that \(A\) is sparse, so \(H\) is sparse. Neither \(S\) nor \(G\) are sparse, but we can still do some sparse matrix computations to solve our problem. In particular, we never want to construct the matrix \(G\) as this may be far too large to fit on a computer!
A course on finite Markov chains will tell us that the Markov chain with transition matrix \(G\) converges to a stationary distribution \(\pi\), i.e. \[\mu^T G^m \to \pi^T,\] for all initial distributions \(\mu\), and in fact this convergence will be quite rapid (certainly if \(\alpha\) is not too close to \(1\) but possibly also otherwise).
So to get an approximation of \(\pi\) it suffices to compute \(\mu G^m\) for a sufficiently large \(m\). This is known as power iteration. How can we do this without ever constructing \(G\)? Consider
\[\begin{align*} \nu^{T}G&=&\mu^{T}\left[\alpha S+(1-\alpha){\bf 1}p^{T}\right]\\ &=&\nu^{T}\left[\alpha H+\alpha dw^{T}+(1-\alpha){\bf 1}p^{T}\right]\\ &=&\alpha\nu^{T}H+\alpha\left(\nu^{T}d\right)w^{T}+(1-\alpha)p^{T}. \end{align*}\]
This suggests computing \(\mu_k = \mu_{k-1}^T G\) for \(k=1,\ldots,m\). The only matrix is the sparse matrix \(H\), and the complexity is \(\mathcal{O}({\rm \#links} + n)\).
Let’s try an example, with a dataset from http://snap.stanford.edu/data/index.html. This is an adjacency matrix with \(875,713\) vertices and \(5,105,039\) edges. The sparse matrix is only 81.7MB, but the whole adjacency matrix would be \(5\) terabytes…
# load the sparse Matrix library
library(Matrix)
# load the web-google data
load("../data/web-google.rdata")
## links[,1] denotes "from" page
## links[,2] denotes "to" page
## note: there are no links from a page to itself
# turn the data into an adjacency matrix
n <- max(links)
numlinks <- dim(links)[1]
# A.sparse is a sparse matrix with A[i,j] = 1 whenever links[k,] = (i,j) for some k
A.sparse <- sparseMatrix(i=links[,1],j=links[,2],x=rep(1,numlinks),dims=c(n,n))
# calculate outlinks where outlinks[i] is the number of outlinks for page i
outlinks <- rep(0,n)
for (i in 1:numlinks) {
outlinks[links[i,1]] <- outlinks[links[i,1]] + 1
}
# calculate d: the binary vector that is 0 when page i has no outlinks
d <- outlinks == 0
# calculate the values to assign to H
# these are nonzero whenever links[k,] = (i,j) for some k but are normalized so that
# sum(A[i,]) = 1 whenever sum(A[i,]) > 0 (i.e. apart from when i is a dangling node)
vsH <- rep(0,numlinks)
for (i in 1:numlinks) {
vsH[i] <- 1/outlinks[links[i,1]]
}
# construct H as a sparse matrix
H.sparse <- sparseMatrix(i=links[,1],j=links[,2],x=vsH,dims=c(n,n))
# set alpha, w and p
alpha <- .85
# these are not sensible choices of w and p, but they "work".
w <- rep(1/n,n)
p <- log(1+(1:n))/sum(log(1+(1:n)))
# we will do power iteration for m iterations
m <- 150
# we will compute the difference between probability vectors after each iteration
diffs <- rep(0,m)
# power iteration
# start off with the uniform distribution
v <- t(rep(1/n,n))
for (i in 1:m) {
v.old <- v # store the old value of v
# compute the new value of v = v.old%*%G
v1 <- alpha*as.vector(v%*%H.sparse)
v2 <- alpha*as.numeric(v%*%d)*t(w)
v3 <- (1-alpha)*t(p)
v <- v1 + v2 + v3
# compute the difference
diff <- sum(abs(v-v.old))
diffs[i] <- diff
# print(paste("iteration ",i,": ||v-v.old||_1 = ",diff,sep=""))
}
diffs [1] 8.511765e-01 3.444636e-01 1.642173e-01 8.847338e-02 5.316488e-02
[6] 3.552487e-02 2.510361e-02 1.857720e-02 1.408584e-02 1.090640e-02
[11] 8.548548e-03 6.784473e-03 5.417833e-03 4.361588e-03 3.527755e-03
[16] 2.868548e-03 2.340552e-03 1.918251e-03 1.575265e-03 1.298393e-03
[21] 1.072019e-03 8.874760e-04 7.357457e-04 6.115093e-04 5.085653e-04
[26] 4.238315e-04 3.535127e-04 2.952880e-04 2.468345e-04 2.066466e-04
[31] 1.730362e-04 1.450712e-04 1.216738e-04 1.021268e-04 8.574776e-05
[36] 7.206109e-05 6.056169e-05 5.093815e-05 4.285723e-05 3.607684e-05
[41] 3.037667e-05 2.559330e-05 2.156369e-05 1.817809e-05 1.532669e-05
[46] 1.292704e-05 1.090422e-05 9.202776e-06 7.767009e-06 6.557833e-06
[51] 5.537997e-06 4.677867e-06 3.951665e-06 3.339420e-06 2.822014e-06
[56] 2.385529e-06 2.016831e-06 1.705447e-06 1.442149e-06 1.219869e-06
[61] 1.031843e-06 8.729511e-07 7.385888e-07 6.250261e-07 5.289005e-07
[66] 4.476567e-07 3.789014e-07 3.207386e-07 2.715203e-07 2.298952e-07
[71] 1.946468e-07 1.648336e-07 1.395896e-07 1.182224e-07 1.001308e-07
[76] 8.481779e-08 7.184392e-08 6.086348e-08 5.156147e-08 4.368410e-08
[81] 3.701155e-08 3.136176e-08 2.657392e-08 2.251987e-08 1.908441e-08
[86] 1.617450e-08 1.370834e-08 1.161918e-08 9.848513e-09 8.348248e-09
[91] 7.076523e-09 5.999015e-09 5.085585e-09 4.311540e-09 3.655360e-09
[96] 3.099217e-09 2.627678e-09 2.228045e-09 1.889193e-09 1.601959e-09
[101] 1.358419e-09 1.151959e-09 9.768745e-10 8.284564e-10 7.025882e-10
[106] 5.958703e-10 5.053714e-10 4.286406e-10 3.635551e-10 3.083738e-10
[111] 2.615645e-10 2.218699e-10 1.882002e-10 1.596487e-10 1.354244e-10
[116] 1.148849e-10 9.745890e-11 8.268000e-11 7.014163e-11 5.950930e-11
[121] 5.048643e-11 4.283410e-11 3.634227e-11 3.083532e-11 2.616301e-11
[126] 2.220031e-11 1.883619e-11 1.598374e-11 1.356204e-11 1.150818e-11
[131] 9.764857e-12 8.287189e-12 7.032220e-12 5.967717e-12 5.064346e-12
[136] 4.298103e-12 3.647556e-12 3.095541e-12 2.627072e-12 2.229465e-12
[141] 1.892181e-12 1.606064e-12 1.362999e-12 1.156989e-12 9.819306e-13
[146] 8.333196e-13 7.073471e-13 6.003143e-13 5.095681e-13 4.325593e-13## rank the pages!
ranking <- sort(v,decreasing=TRUE,index.return=TRUE)$ix3 Parallel computing
Traditionally, computing was largely serial. By this we mean that, outside certain exceptional cases, algorithms were typically designed to be run on a single machine with one processor on one thread. Of course, general purpose computers have operating systems that allowed several programs to run simultaneously using a single processor: think of a personal computer with a graphical interface and several different open programs. These only gave the impression of running simultaneously; the operating system switches between tasks in such a way that humans rarely notice the difference.
Since 2005 or so, however, there has been a shift to larger number of cores per processor and processors per computer. This is due to physical difficulties associated with making individual cores more powerful. Designing algorithms that run fast on modern computers sometimes requires utilizing several cores simultaneously, which necessitates splitting computation into parts that can run in parallel.
Graphical processing units take this to one extreme, with very large numbers of cores and relatively less hardware devoted to control flow. This means there can be very significant speedups for very specific types of parallel computation.
It is useful to distinguish between different types of parallel computation. First we need to understand at a high level how computers compute. A rough description is:
- A processor may have several cores.
- A core can execute instructions and access various forms of memory, usually arranged in a hierarchy (fast to slow: caches, main memory, disk).
- A thread (of execution) is a sequence of instructions to be run on a core.
- Multiple threads can be part of the same process, and can thereby share resources with each other.
- Distinct processes do not share resources (at least directly).
Now for different modes of parallelism:
- Running several processes in parallel (on one machine or several).
- Running several threads in parallel within a process (on one machine).
In the first case, one may have machines connected via a network and allow messages to be passed between them. Some would call this distributed computing.
In a simple parallel computing scenario, you have lots of numbers to crunch that have no dependence (i.e. there is no need to compute them in any specific order), so you can just run the relevant programs on the same or different machines. This is an easy win, and tends to give a factor speedup that is roughly the number of cores available. One caveat is that the programs may all require lots of memory (e.g. to load a big dataset) and if you construct several processes the memory available may be exhausted. In such cases one may want to look at threads.
In more complicated scenarios, there is a lot of dependence. For example, you may need to compute lots of numbers in a for loop that are all required for the subsequent iteration of an iterative algorithm. For this it makes sense to have several threads (which share memory) each do some of the computations. This requires writing programs that are multi-threaded.
For some hardware architectures, like GPUs, there are a lot of cores but (of course it gets a bit technical and we can’t go into the details) but blocks of computation can only take place perfectly in parallel if all of the computations are the same, down to the instructions. This is very different to just running different processes on different cores, and requires much finer attention to detail. In practice, people often used frameworks with domain-specific languages that enable the computation to be coerced into a suitable form.
One final thing: memory management becomes incredibly important when moving from high-level interpreted languages like R or Python to lower-level instructions as, e.g., produced by languages like C or Julia. This is important for parallel computing and serial computing alike.
To get a taste, consider the rough speed of different types of memory access:
- L1 cache (around 64KB) reference: 0.5ns.
- L2 cache (around 256KB) reference: 7ns.
- Main memory (around 4–8GB) reference: 100ns.
- Disk seek: 10ms = \(10^7\)ns.
- Solid State Drive: 0.1ms = \(10^5\) ns.
By analogy:
- L1 cache: reaching for something on our desk (e.g. 1 second)
- L2 cache: fetching it instead from a drawer (14 seconds)
- Main memory: going up a set of stairs into another room to fetch something (3 minutes, 20 seconds).
- Disk: walking from the University of Warwick to Cape Town (South Africa) and back (5555 hours or 231 days).
- Solid state drive: walk to Brighton (55.55 hours or 2.31 days).
This makes clear that the simple model we often use to talk about algorithmic complexity (e.g. every operation takes 1 unit of time) is not accurate for certain types of computation. In particular, it ignores the importance of memory management and this tends to be very important for optimizing computations that are used very often.
4 Random algorithms
Random algorithms are algorithms that output (realizations of) random variables. That is, the output for a given fixed output is random. This is different to “randomized” algorithms that utilize randomness but whose output is not random, e.g. randomized quicksort.
Technically, a random algorithm is just an algorithm with an additional random input and whose output can correspondingly depend on the random input.
There are many open questions about the complexity of random vs. deterministic algorithms.
A very cute example of a random algorithm is Freivald’s algorithm for checking matrix multiplication. Say we have as input 3 \(n \times n\) matrices, \(A\), \(B\) and \(C\). We desire an algorithm that outputs “yes” if \(A \times B = C\) and “no” otherwise. The standard deterministic approach is to just do the multiplication \(A \times B\) in \(O(n^\alpha)\) time, where \(\alpha = 2.373\) corresponds to the best matrix multiplication complexity we know of, and check if the result is equal to \(C\).
If we are willing to be wrong with a very small probability, we can use a random algorithm that will always return “yes” if \(A \times B = C\) and will return “no” with probability \(1-2^{-k}\) if \(A \times B \neq C\), where the complexity of the algorithm is \(O(kn^2)\). We’ll see how it works by considering one random check: the idea is to
- sample a binary vector \(\xi\) valued in \(\{0,1\}^n\) such that each component is a \({\rm Bernoulli}(1/2)\) random variable.
- Compute \(Y = A(B\xi)\) and \(Z = C\xi\), which takes \(O(n^2)\) time as it’s matrix-vector multiplication.
- If \(AB = C\), then \(Y = Z\).
- If \(Y \neq Z\), then \(AB \neq C\).
The main analysis to do is to show that if \(AB \neq C\) then the probability that \(Y \neq Z\) is at least \(1/2\). So, assume \(AB \neq C\). Then, consider \(R = (AB-C)\xi = D\xi\), where \(D = AB-C\). We are checking whether \(R\) has a non-zero element.
Let \(d_{ij}\) be a nonzero element of \(D\), which exists because \(AB \neq C\). Then consider \[R_i = \sum_{k=1}^n d_{ik}\xi_k = d_{ij}\xi_j + \sum_{k=1,k\neq j}^n d_{ik}\xi_k = d_{ij}\xi_j + W,\] Then, \[\mathbb{P}(R_i=0) = \mathbb{P}(R_i=0\mid W=0)\mathbb{P}(W=0) + \mathbb{P}(R_i=0\mid W\neq 0)\mathbb{P}(W \neq 0).\] We find \[\mathbb{P}(R_i=0\mid W=0) = \mathbb{P}(\xi_j=0) = \frac{1}{2},\] and \[\mathbb{P}(R_i=0\mid W\neq 0) \leq \mathbb{P}(\xi_j=1) = \frac{1}{2},\] so \[\mathbb{P}(R_i=0) \leq \frac{1}{2} \left [ \mathbb{P}(W=0) + \mathbb{P}(W\neq 0) \right ] = \frac{1}{2}.\] For an algorithm with the desired probability of correctness, we can just repeat the procedure \(k\) times, outputting “no” if any of the trials result in \(Y \neq Z\) and outputting “yes” if \(Y=Z\) in all of the trials. Again, the only possible error is that \(AB \neq C\) but we output “yes”: the probability of this occurring is less than or equal to \(2^{-k}\).
5 Hashing
Suppose that you want to store data, which is to be accessed using arbitrary keys. For example, you want to store data on a large number of people, using their names (as a text string) as the key for finding the record in memory. i.e., data about someone is read in from somewhere, and is stored at a location in computer memory, depending on what their name string is. Later you want to retrieve this information using their name string. How can you actually do this?
You could store some sort of directory of names, which would be searched through to find the location of the data. But such a search is always going to take some time, which will increase with the number of people’s data that is stored. Can we do better? With hashing the answer is yes. The pivotal idea is to create a function which will turn an arbitrary key into an index which is either unique or shared with only a few other keys, and has a modest predefined range. This index gives a location in a (hash) table which in turn stores the location of the actual data (plus some information for resolving `clashes’). So given a key, we use the (hash) function to get its index, and its index to look up the data location, with no, or very little searching. The main problem is how to create a hash function.
An elegant general strategy is to use the ideas of pseudorandom number generation to turn keys into random integers. For example, if the key were some 64bit quantity, we could use it to seed the Xorshift random number generator given earlier, and run this forward for a few steps. More generally we would need an approach that successively incorporated all the bits in a key into the `random number’ generation process (see e.g. section 7.6 of Press et al., 2007, for example code). So, the hash function should return numbers that appear random, which will mean that they tend to be nicely spread out, irrespective of what the keys are like. Of course, despite this, the function always returns the same number for a given key.
It remains to turn the number returned by the hash function, \(h\), into an index, \(i_h\) , somewhere between 0 and the length, \(n_h\), of the hash table minus 1. \(i_h = h~ {\rm mod}~ n_h\) is the usual approach. At position \(i_h\) in the hash table we might store the small number of actual keys (or possibly just their \(h\) values) sharing the \(i_h\) value, and the actual location in memory of the corresponding data.
The really neat thing here is that the randomness of the \(h\)’s means that whatever the keys look like, you usually end up with a fairly uniform distribution of \(i_h\)’s, and only a small number of keys sharing each slot in the hash table. It is this that makes data retrieval so efficient, relative to searching a directory of keys.
Note that R environments are themselves methods for associating values with lookup keys, and in fact, R environments can use hash tables: see ?new.env.
ht <- new.env(hash=TRUE)
ht[["foo"]] = "bar"
ht[["baz"]] = 1:10
ht[["foo"]][1] "bar"ht$baz [1] 1 2 3 4 5 6 7 8 9 10If that doesn’t sound like it has any application in statistics, consider the construction of triangulations in spatial statistics (a triangulation being a way of joining a set of points up with triangles, in such a way that the whole convex hull of the points is covered in triangles). Efficient construction of triangulations is tricky, and several efficient algorithms work by incrementally adding triangles. You usually know at least one neighbour of each triangle at its creation, but you actually need to know all its neighbours in the existing set. The task of finding those neighbours would be prohibitive if you had to search each existing triangle for edges shared with the new triangle. Instead one can maintain a hash table for unpaired triangle edges, where the hash key is based on the end co-ordinates of each edge. This allows any newly created edge to be paired with any existing edge in little more than the length of time it takes to create the hash key.
6 Further reading
Gentle, J.E. (2003) Random Number Generation and Monte Carlo Methods (2nd ed), Springer, Chapters 1 and 2 provides up to date summaries both of uniform generators, and of generator testing. Chapter 3 covers Quasi Monte Carlo, and the remaining chapters cover simulation from other distributions + software.
Matsumoto, M. and Nishimura, T. (1998) Mersenne Twister: A 623-dimensionally equidistributed uniform pseudo-random number generator, ACM Transactions on Modeling and Computer Simulation, 8, 3-30. This gives details and code for the Mersenne Twister.
http://www.math.sci.hiroshima-u.ac.jp/~m-mat/MT/emt.html gives the Mersenne Twister source code.
Marsaglia, G. (2003) Xorshift RNGs, Journal of Statistical Software, 8(14):1-6.
DIEHARD is George Marsaglia’s Diehard battery of tests for random number generators
Press, W.H., S.A. Teukolsky, W.T. Vetterling and B.P Flannery (2007) Numerical Recipes (3rd ed.) Cambridge. Chapter 7, on random numbers is good and reasonably up to date, but in this case you do need the third edition, rather than the second (and certainly not the first).
Ripley (1987, re-issued 2006) Stochastic Simulation, Wiley. Chapter’s 2 and 3 are where to go to really get to grips with the issues surrounding random number generation. The illustrations alone are an eye opener. You probably would not want to use any of the actual generators discussed and the remarks about mixed generators now look dated, but the given principles for choosing generators remain sound (albeit with some updating of what counts as simple, and the integers involved). See chapter 3 and 4 for simulating from other distributions, given uniform deviates.
Gamerman, D. and H.F. Lopes (2006) Markov Chain Monte Carlo: Stochastic simulation for Bayesian inference, CRC. Chapter 1 has a nice compact introduction to simulation, given i.i.d. \(U(0,1)\) deviates.
Devroye, L., (2003) Non-Uniform Random Variate Generation, Springer. This is a classic reference. Now out of print.
Steele Jr, Guy L., Doug Lea, and Christine H. Flood. “Fast splittable pseudorandom number generators.” ACM SIGPLAN Notices 49.10 (2014): 453-472. In the context of parallel computing, sequential random number generators are very problematic. Splittable generators turn out to be very useful in this context.
For a good introduction to the sparse methods underlying Martin Maechler and Douglas Bates’ R package
Matrix, see Davis, T. A. (2006) Direct Methods for Sparse Linear Systems, SIAM.Tim Davis’s home page is worth a look for more recent sparse matrix developments.
If you think hashing is clever, take a look at the rest of Cormen, T., C.E. Leiserson, R.L. Rivest and C. Stein (2001) Introduction to Algorithms (2nd ed.) MIT.
Once again, Press, W.H., S.A. Teukolsky, W.T. Vetterling and B.P Flannery (2007) Numerical Recipes (3rd ed.) provides a readable introduction to a much wider range of topics in scientific computing than has been covered here.